Karyotype complexity is established by enumerating the number of chromosome abnormalities present in a karyotype. In chronic lymphocytic leukemia (CLL), karyotype complexity, defined as ≥3 chromosome abnormalities, is an important prognostic indicator associated with poor outcomes. Additionally, stratifying patients (pts) by highly complex karyotypes (≥5 chromosome abnormalities) has shown significantly poorer outcomes compared to pts with 3-4 abnormalities (Baliakas, Blood, 2019). In principle, counting chromosome abnormalities is straightforward; however, in practice it can be nuanced, and the number of abnormalities counted for the same karyotype can differ across laboratories. A lack of standardized rules has resulted in variability in establishing complexity across CLL studies. We and others have proposed adapting the international working group on myelodysplastic syndrome (MDS) framework for use in CLL (Chun, Leuk Res, 2010; Jondreville, Am J Hematol, 2020). Additionally, a separate set of guidelines were published in the 2020 International System for Human Cytogenomic Nomenclature (ISCN). To investigate how different guidelines may impact patient categorization in correlative studies, we evaluated karyotype complexity using both guidelines in a previously reported, well-characterized, dataset with long term follow-up of patients with CLL treated with ibrutinib (Kittai, Blood, 2021).
456 patient karyotypes underwent expert review by two cytogeneticists. Discordant scores were discussed to reach consensus. Pts were categorized as not complex (0-2 abnormalities), complex (3-4), or highly complex (≥5) for each set of guidelines. The weighted kappa was used to evaluate agreement between the two guidelines. Progression free survival (PFS) from ibrutinib start and overall survival (OS) were evaluated by the Kaplan-Meier method. Univariable Cox regression model discrimination power for PFS and OS were evaluated by Uno's concordance statistic.
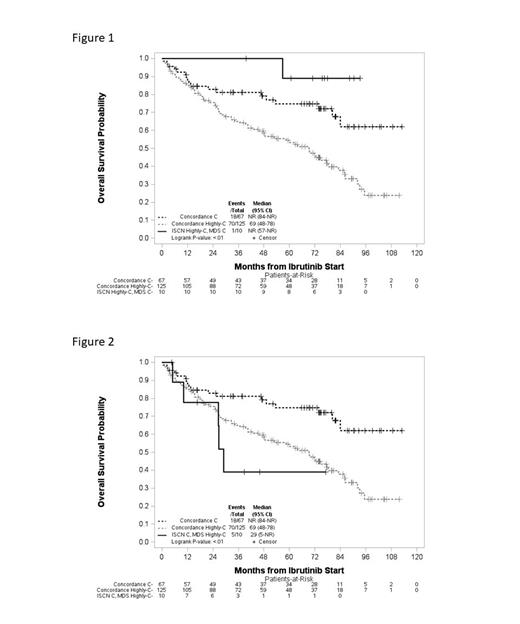
MDS guidelines categorized 50.2% as not complex, 19.7% complex and 30.1% as highly complex; similarly, ISCN categorized 50.2% as not complex, 20.2% complex and 29.6% as highly complex. Complexity categories assigned by the two guidelines had a weighted kappa agreement of 0.88 (95% CI 0.84-0.91), with 11% of pts placed in discordant complexity categories. Albeit numbers of pts were small, evaluation of these discordant cases compared to the consensus concordant cases showed outcomes varied. Discordant cases categorized by the MDS guidelines as complex and by ISCN as highly complex showed longer OS compared to the concordant complex and highly complex cases, suggesting ISCN overrated complexity (Fig 1). Conversely, discordant cases categorized by MDS guidelines as highly complex while ISCN categorized as complex showed decreased OS compared to the concordant complex and highly complex cases, suggesting ISCN underrated complexity in these cases (Fig 2). PFS trended similarly for these groups.
Both the MDS and the ISCN guidelines showed increased karyotype complexity was significantly associated with shorter PFS (both with p<0.0001) and OS (both with p<0.0001). Comparing the classifiers not complex, complex and highly complex for the entire cohort found no significant difference between the prediction power of the MDS and ISCN guidelines for PFS. For OS, the MDS guidelines had a statistically stronger prediction power compared to ISCN (p=0.0235). Further, when the poorest prognostic group (highly complex) was compared to all others, the MDS guidelines showed significantly stronger prediction power for both PFS (p=0.0359) and OS (p=0.0039) compared to ISCN.
Although both sets of guidelines were prognostically significant, in this predominately relapsed/refractory CLL patient population receiving ibrutinib, the MDS guidelines performed more robustly when used as a categorical variable. Clinical trials incorporating karyotype complexity for prognostication, or for treatment stratification, should specify a set of guidelines to apply consistently across the trial. Given the importance of karyotype complexity, particularly with targeted therapies, further studies are needed to evaluate these two sets of guidelines across varying CLL populations to establish recommendations for assigning karyotype complexity uniformly in CLL.
Disclosures
Miller:AbbVie: Research Funding. Kittai:Eli Lilly: Consultancy; BeiGene: Consultancy, Research Funding, Speakers Bureau; AstraZeneca: Consultancy, Research Funding; Abbive: Consultancy; Janssen: Consultancy; KITE: Consultancy; BMS: Consultancy. Bhat:Aptitude Health: Honoraria; Abbvie: Consultancy; AstraZeneca: Consultancy, Research Funding. Rogers:Janssen: Consultancy; Loxo@Lilly: Consultancy; Novartis: Research Funding; Beigene: Consultancy; AstraZeneca: Consultancy; Pharmacyclics: Consultancy; Genentech: Consultancy, Research Funding; AbbVie: Consultancy, Research Funding. Woyach:Newave: Consultancy; Loxo: Consultancy; Beigene: Consultancy; AstraZeneca: Consultancy; Abbvie: Consultancy; Schrodinger: Research Funding; Morphosys: Research Funding; Karyopharm: Research Funding; Janssen: Consultancy, Research Funding; Pharmacyclics: Consultancy, Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal